Current state-of-the-art methods for object detection rely on annotated bounding boxes of large data sets for training. However, obtaining such annotations is expensive and can require up to hundreds of hours of manual labor. This poses a challenge, especially since such annotations can only be provided by experts, as they require knowledge about the scientific domain. To tackle this challenge, we propose a domain-specific weakly supervised object detection algorithm that only relies on image-level annotations, which are significantly easier to acquire. Our method distills the knowledge of a pre-trained model, on the task of predicting the presence or absence of a virus in an image, to obtain a set of pseudo-labels that can be used to later train a state-of-the-art object detection model. To do so, we use an optimization approach with a shrinking receptive field to extract virus particles directly without specific network architectures. Through a set of extensive studies, we show how the proposed pseudo-labels are easier to obtain, and, more importantly, are able to outperform other existing weak labeling methods, and even ground truth labels, in cases where the time to obtain the annotation is limited.
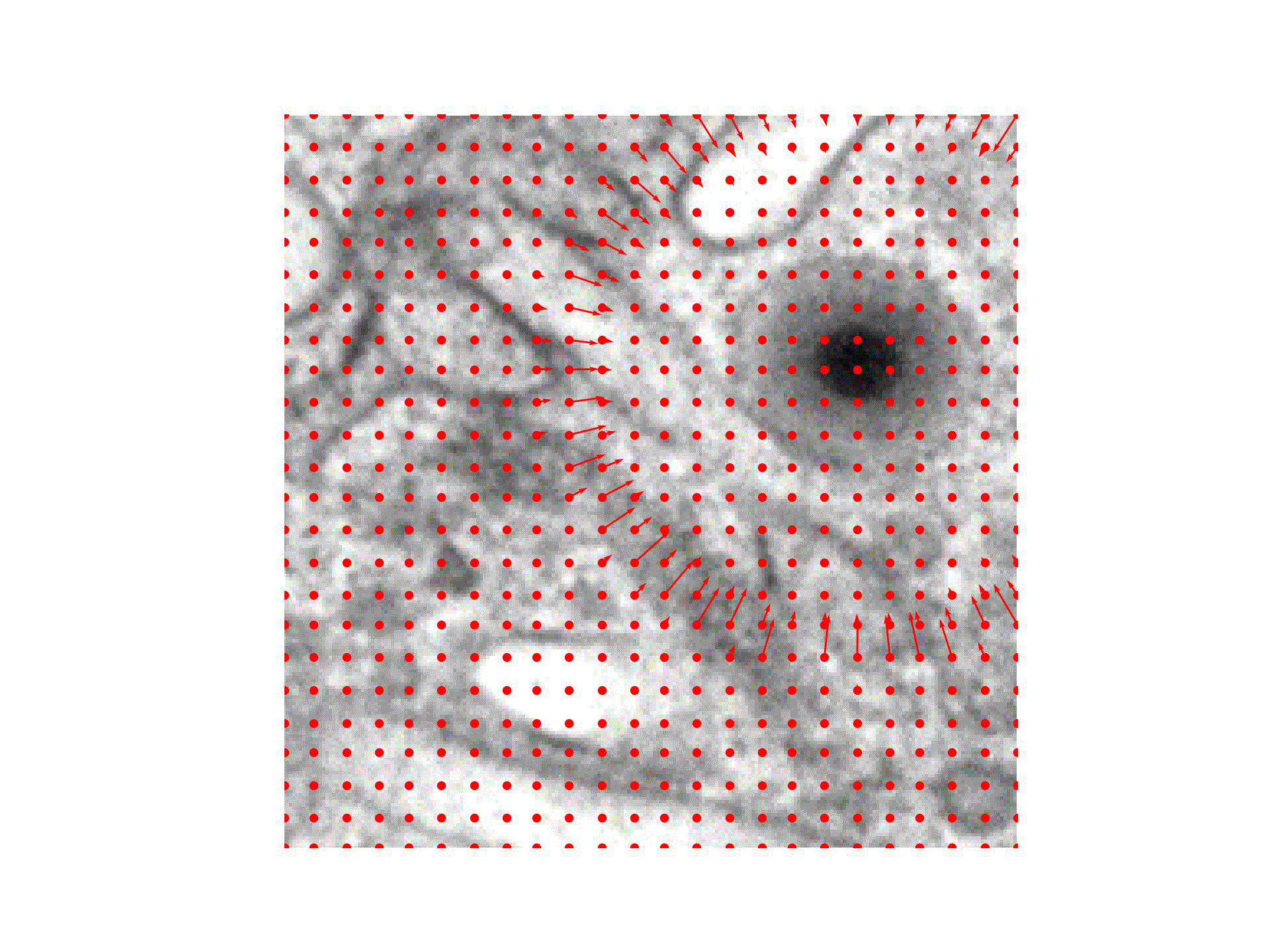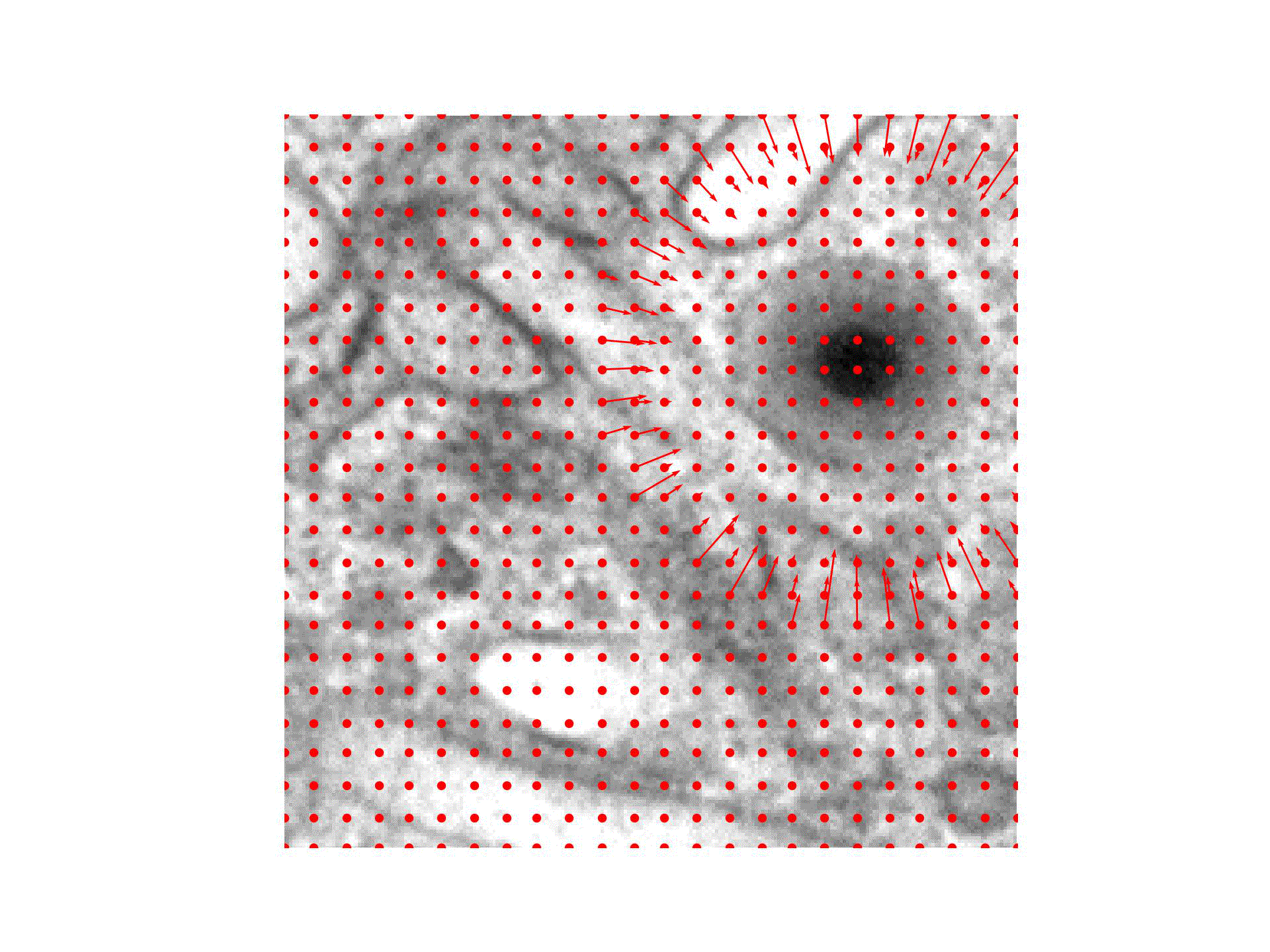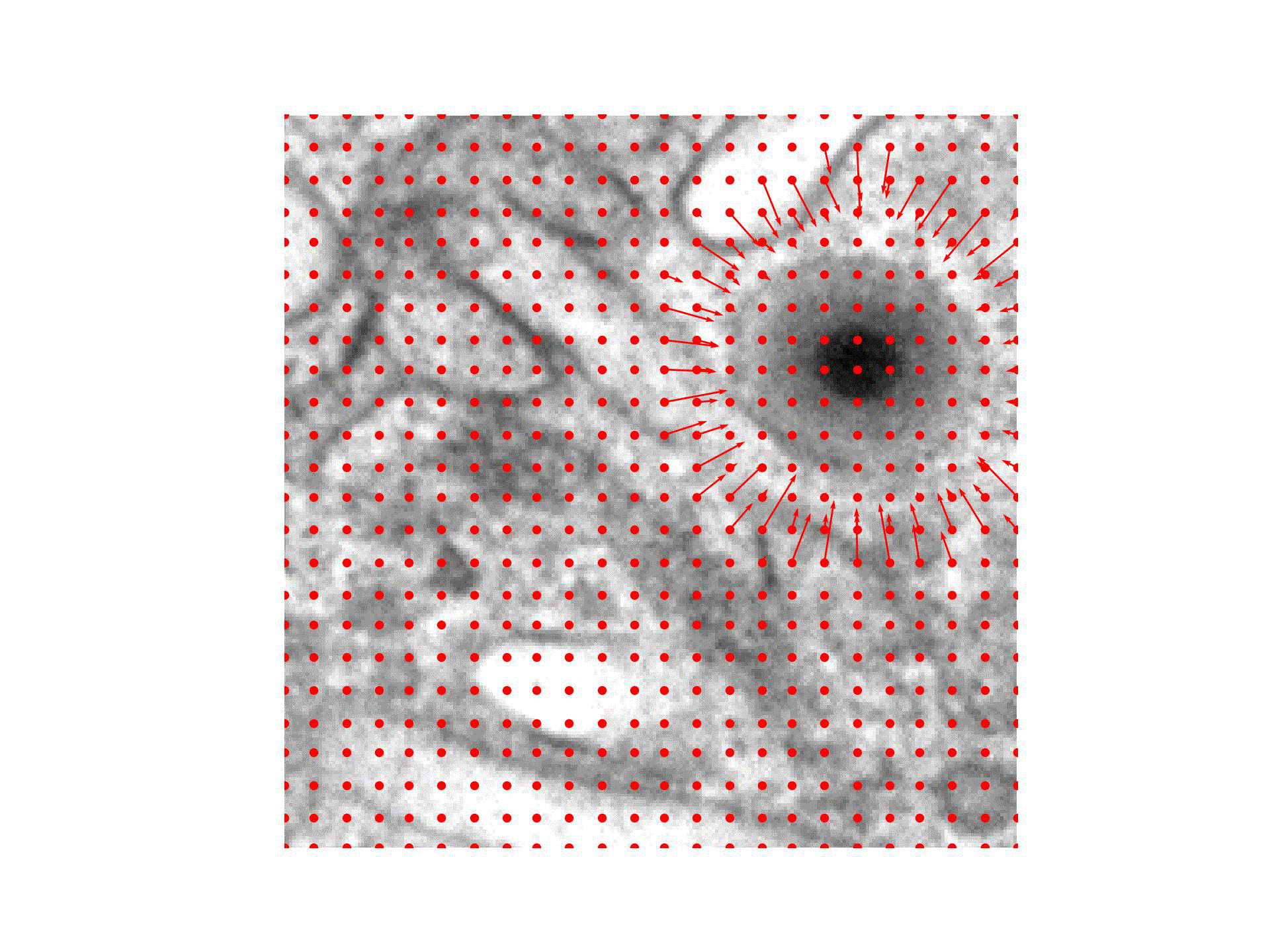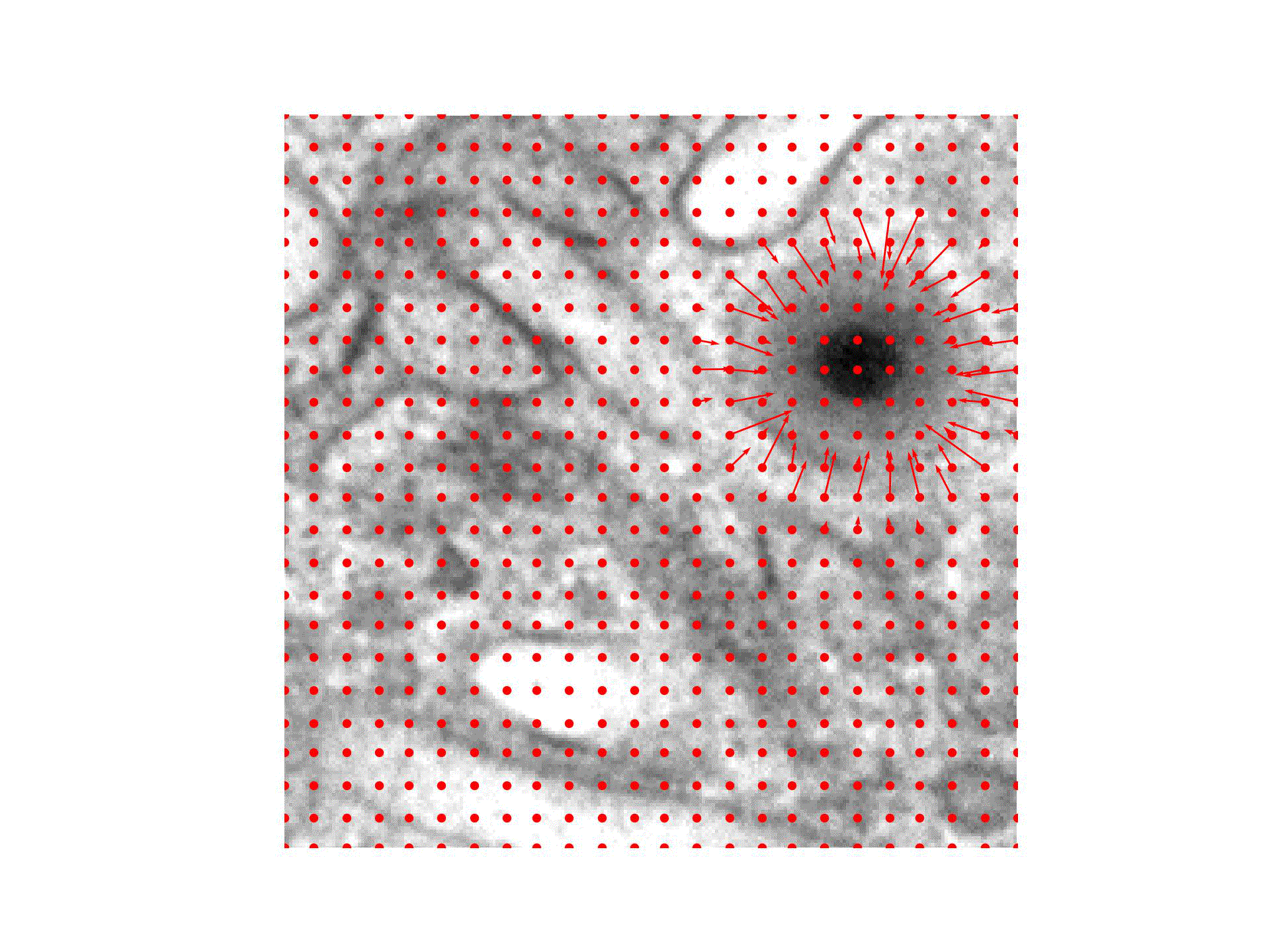
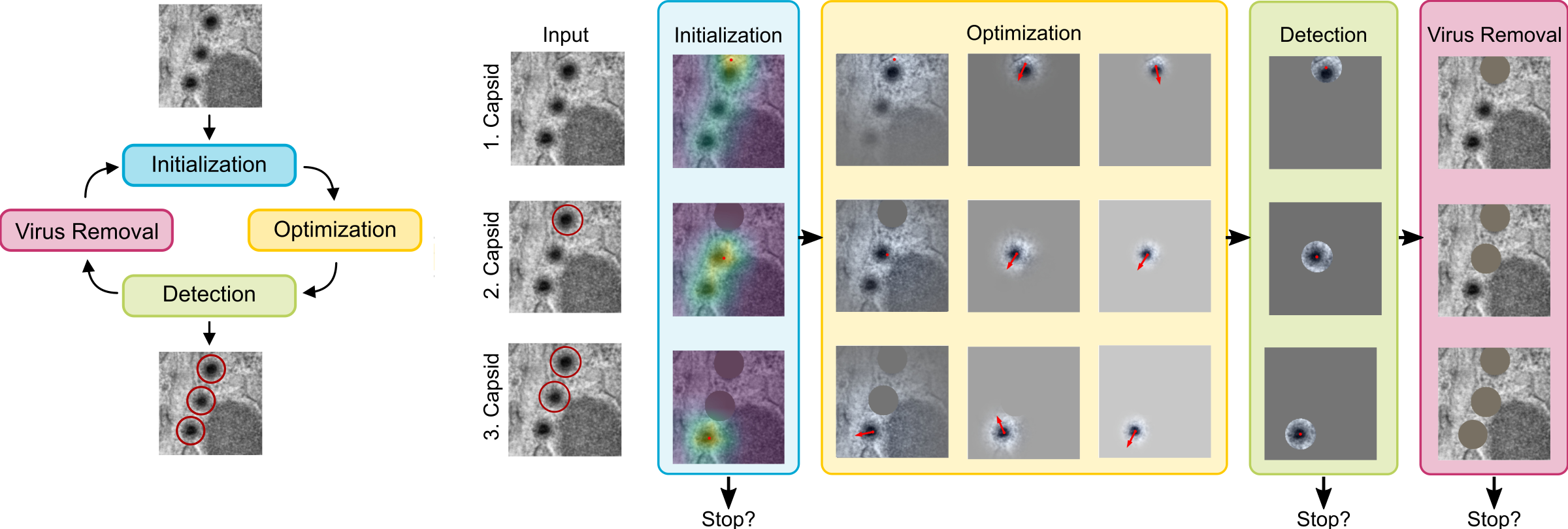 We are introducing an iterative optimization process with shrinking receptive field to generate accurate bounding boxes for virus detection.
In case of virus capsid detection, one can approximate the size of the bounding box from single instances, as the virus size does not vary.
Additionally, the sizes are usually known and can hence be derived from literature.
Our approach therefore needs to find the center of the virus capsid to be able to detect it.
We hence train a classifier to predict the presence of virus capsids in the input image and reuse it during the optimization:
For the Initialization of the particle position we compute a CAM obtained through GradCAM and place it at the position of the highest CAM value.
During Optimization the position is iteratively refined, guided by the pre-trained classifier output and a Gaussian mask with decreasing standard deviation centered at the current position.
A Detection is happening once the position converged to the exact position of the virus particle.
Finally, the input image is prepared to detect the next virus by the Virus Removal of previously detected virus particles.
We check at multiple points of the virus detection pipeline, if a stopping criteria is met.
The collected bounding boxes can be directly applied (Ours(Opt)) or further used to train an object detection model (Ours(OD)).
We are introducing an iterative optimization process with shrinking receptive field to generate accurate bounding boxes for virus detection.
In case of virus capsid detection, one can approximate the size of the bounding box from single instances, as the virus size does not vary.
Additionally, the sizes are usually known and can hence be derived from literature.
Our approach therefore needs to find the center of the virus capsid to be able to detect it.
We hence train a classifier to predict the presence of virus capsids in the input image and reuse it during the optimization:
For the Initialization of the particle position we compute a CAM obtained through GradCAM and place it at the position of the highest CAM value.
During Optimization the position is iteratively refined, guided by the pre-trained classifier output and a Gaussian mask with decreasing standard deviation centered at the current position.
A Detection is happening once the position converged to the exact position of the virus particle.
Finally, the input image is prepared to detect the next virus by the Virus Removal of previously detected virus particles.
We check at multiple points of the virus detection pipeline, if a stopping criteria is met.
The collected bounding boxes can be directly applied (Ours(Opt)) or further used to train an object detection model (Ours(OD)).
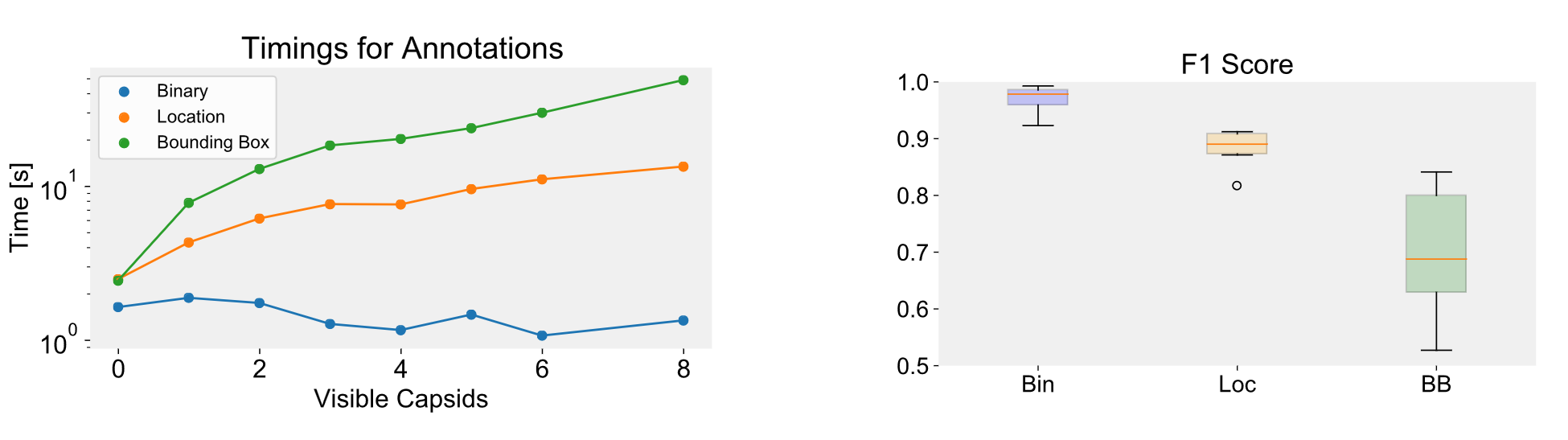 Our user study reveals that using bounding boxes and location labels during annotation leads to additional workload and higher error rates,
resulting in increased annotation time compared to image-level labels. Image-level labels are found to be less error-prone and allow for faster annotation.
Our user study reveals that using bounding boxes and location labels during annotation leads to additional workload and higher error rates,
resulting in increased annotation time compared to image-level labels. Image-level labels are found to be less error-prone and allow for faster annotation.
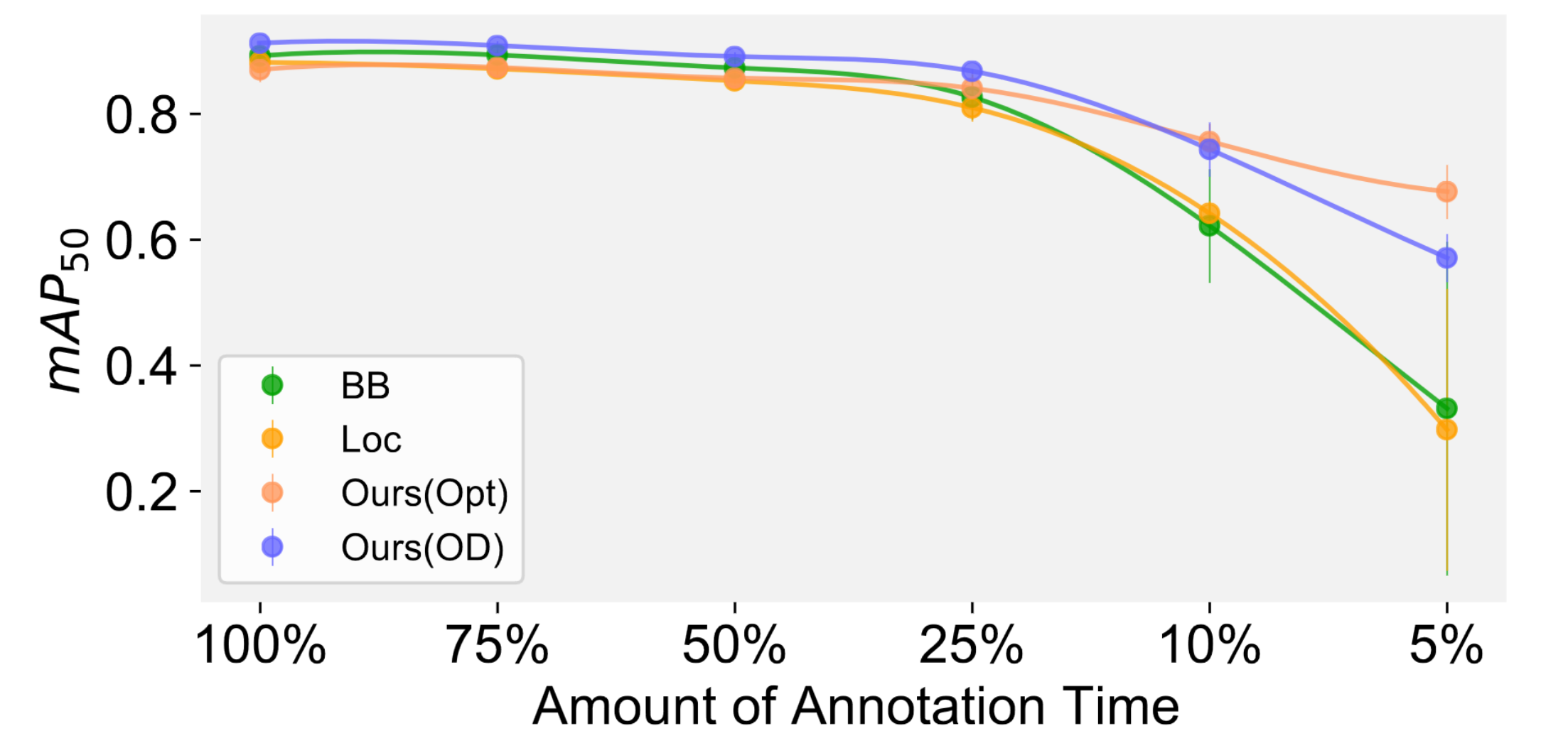 We hence compare the performance of detecting virus capsids using bounding box labels, location labels and our approach using image level labels when the annotation time is fixed.
We found that when the annotation time is limited, our approach is able to outperform location as well as ground truth labels.
We hence compare the performance of detecting virus capsids using bounding box labels, location labels and our approach using image level labels when the annotation time is fixed.
We found that when the annotation time is limited, our approach is able to outperform location as well as ground truth labels.
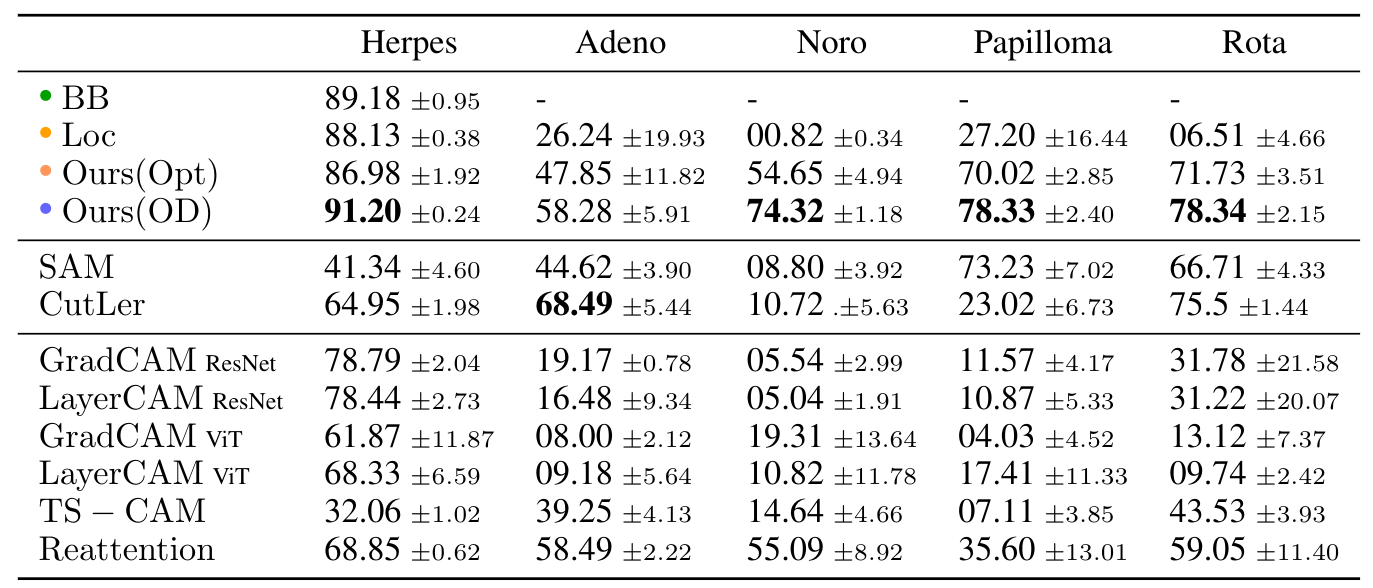 Most state-of-the-art weakly supervised approaches thrive on large dataset sizes with object centric nature.
We hence found that all compared approaches were not able to outperform our approach for the detection of virus capsids in EM images.
We here compare against two adapted zero shot methods: SAM and CutLer, as well as multiple weakly supervised approaches.
For a fair comparison we also include the virus size in the existing approaches.
Most state-of-the-art weakly supervised approaches thrive on large dataset sizes with object centric nature.
We hence found that all compared approaches were not able to outperform our approach for the detection of virus capsids in EM images.
We here compare against two adapted zero shot methods: SAM and CutLer, as well as multiple weakly supervised approaches.
For a fair comparison we also include the virus size in the existing approaches.
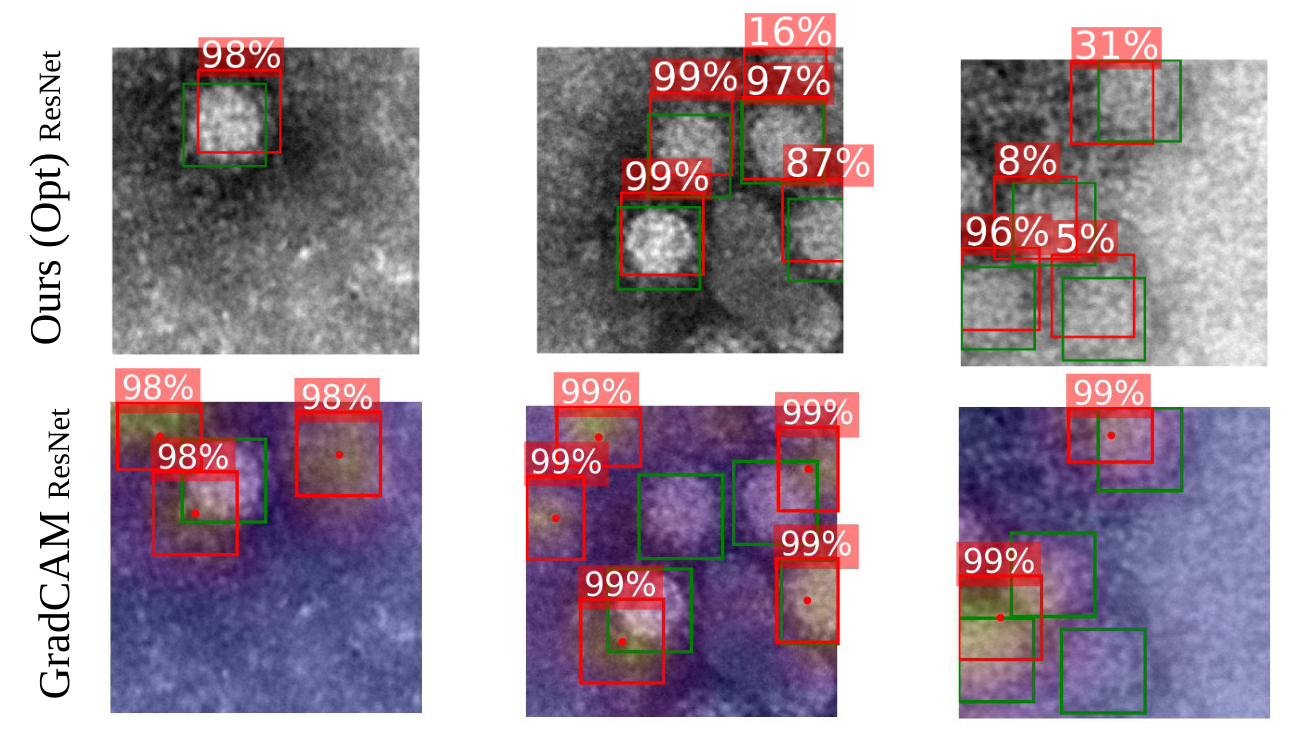 Even though there is a clear bias visible towards the boarder of the virus (which is most
likely due to the imaging modality of negative stain TEM), our approach is able to converge to suitable
positions of the virus based on the introduced optimization strategy. The GradCAM approach, which
was applied to the same classifier, on the other hand, is not able to produce well fitting bounding
boxes.
Even though there is a clear bias visible towards the boarder of the virus (which is most
likely due to the imaging modality of negative stain TEM), our approach is able to converge to suitable
positions of the virus based on the introduced optimization strategy. The GradCAM approach, which
was applied to the same classifier, on the other hand, is not able to produce well fitting bounding
boxes.
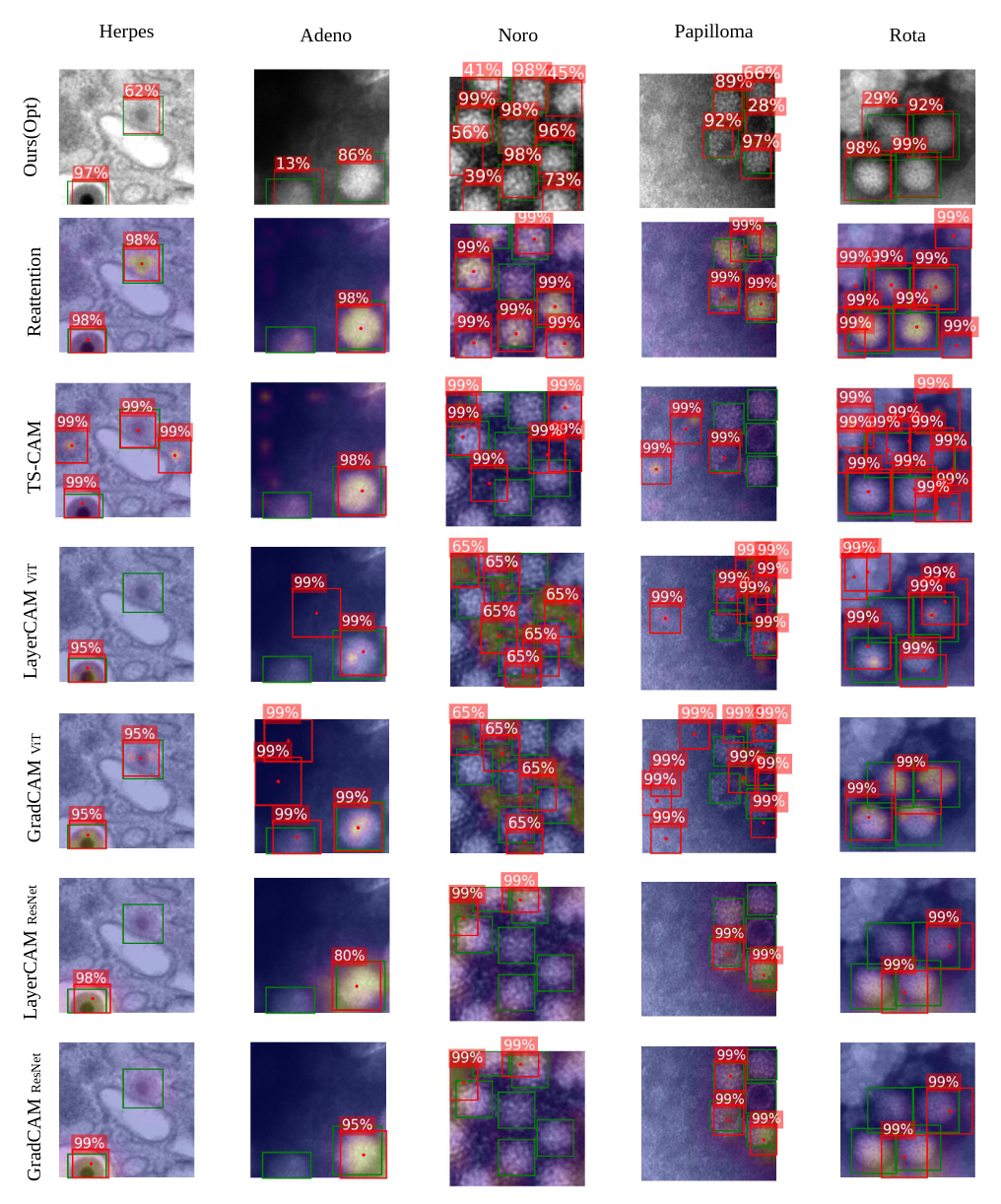 Qualitative results showing Ours(Opt) and other weakly supervised approaches. Note that other methods mainly fail when there are nore instances of the virus visible in the input image.
Additionally, other approaches are prone to detect noise as virus particles.
Qualitative results showing Ours(Opt) and other weakly supervised approaches. Note that other methods mainly fail when there are nore instances of the virus visible in the input image.
Additionally, other approaches are prone to detect noise as virus particles.
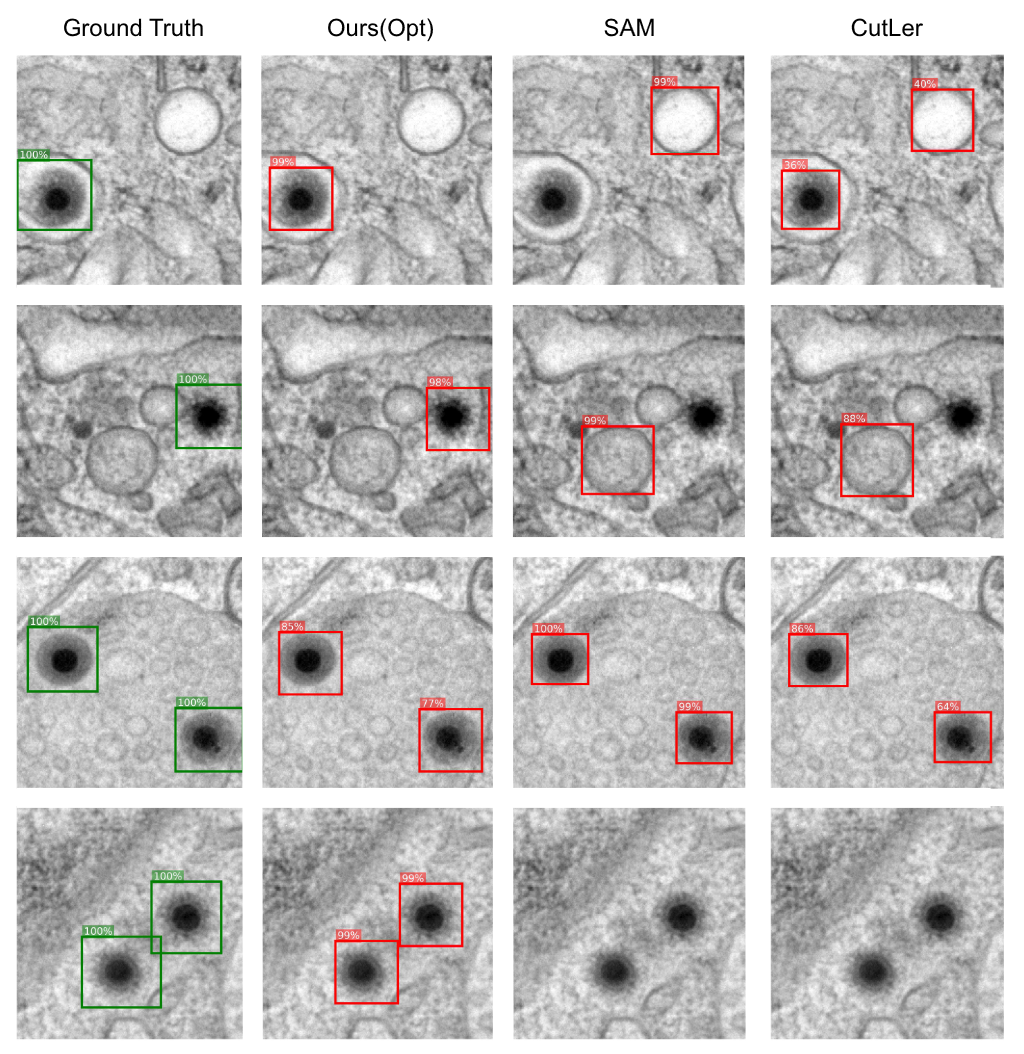 Qualitative results on the zero shot methods show that they fail to detect naked virus particles, that are not clearly separated from their surroundings. Additionally, other objects, that are similar in size as the virus are detected as capsids.
This is due to the zero shot application of these methods.
However, retraining the methods is not trivial, as SAM requires large datasets and annotations and CutLer relies on a large scale pretraining dataset which is not trivial to collect in the case of EM, as sample preparation and imageing is time and cost consuming.
Qualitative results on the zero shot methods show that they fail to detect naked virus particles, that are not clearly separated from their surroundings. Additionally, other objects, that are similar in size as the virus are detected as capsids.
This is due to the zero shot application of these methods.
However, retraining the methods is not trivial, as SAM requires large datasets and annotations and CutLer relies on a large scale pretraining dataset which is not trivial to collect in the case of EM, as sample preparation and imageing is time and cost consuming.
@inproceedings{kniesel2024weakly,
title={Weakly Supervised Virus Capsid Detection with Image-Level Annotations in Electron Microscopy Images},
author={Kniesel, Hannah and Sick, Leon and Payer, Tristan and Bergner, Tim and Shaga Devan, Kavitha and Read, Clarissa and Walther, Paul and Ropinski, Timo and Hermosilla, Pedro},
booktitle={Proceedings of International Conference on Learning Representations}
year={2024}
}